It’s not all Spike! 👉Emergence of novel subgenomic mRNAs in SARS-CoV-2. Sharing our latest preprint with work led by @Artisplicer, showing effectively the coronavirus analogue of entirely new promoters and splice sites emerging in the genome 🆒 biorxiv.org🧵 1/n
This process is a hallmark of coronavirus evolution and adaptation to new hosts, and we are seeing it in real-time… It’s also reminiscent of eukaryotic gene evolution via exon shuffling and duplication of promoter and regulatory elements. 2/n
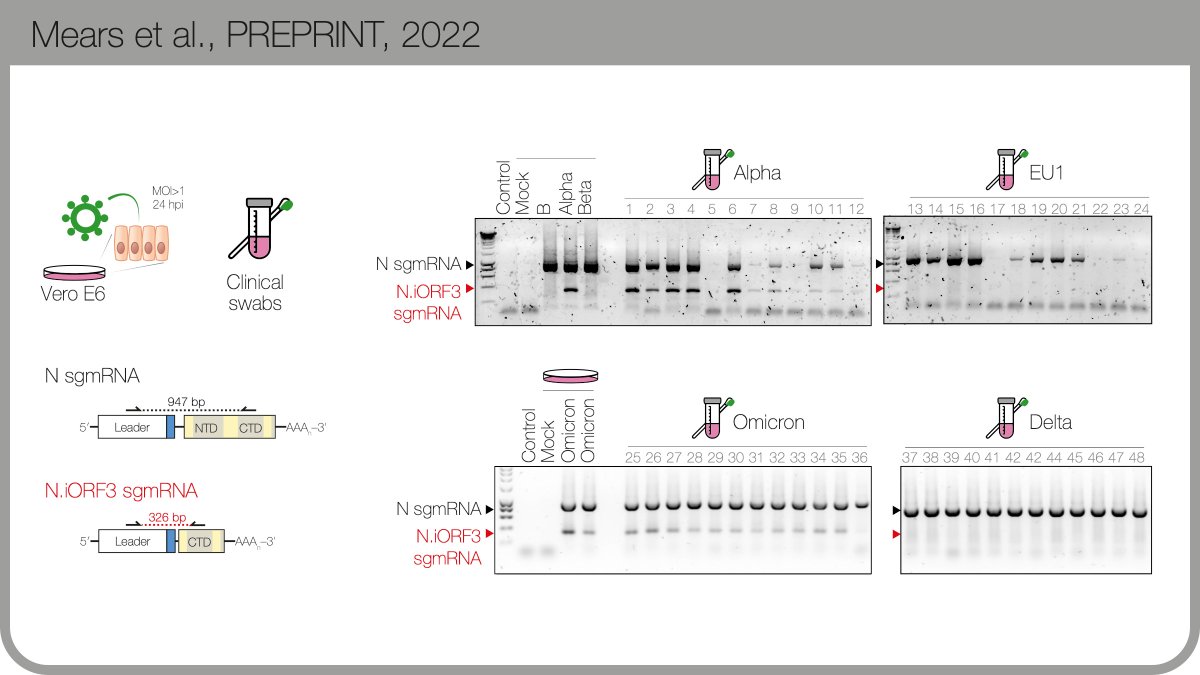
This new sgmRNA is expressed in culture & patient samples, available from the @TheCrick + @UCLHResearch Legacy Study. Careful RT-qPCR quantitation suggests there’s as much of this new subgenomic mRNA (sgmRNA) as there is canonical viral “E” gene sgmRNA. 4/n
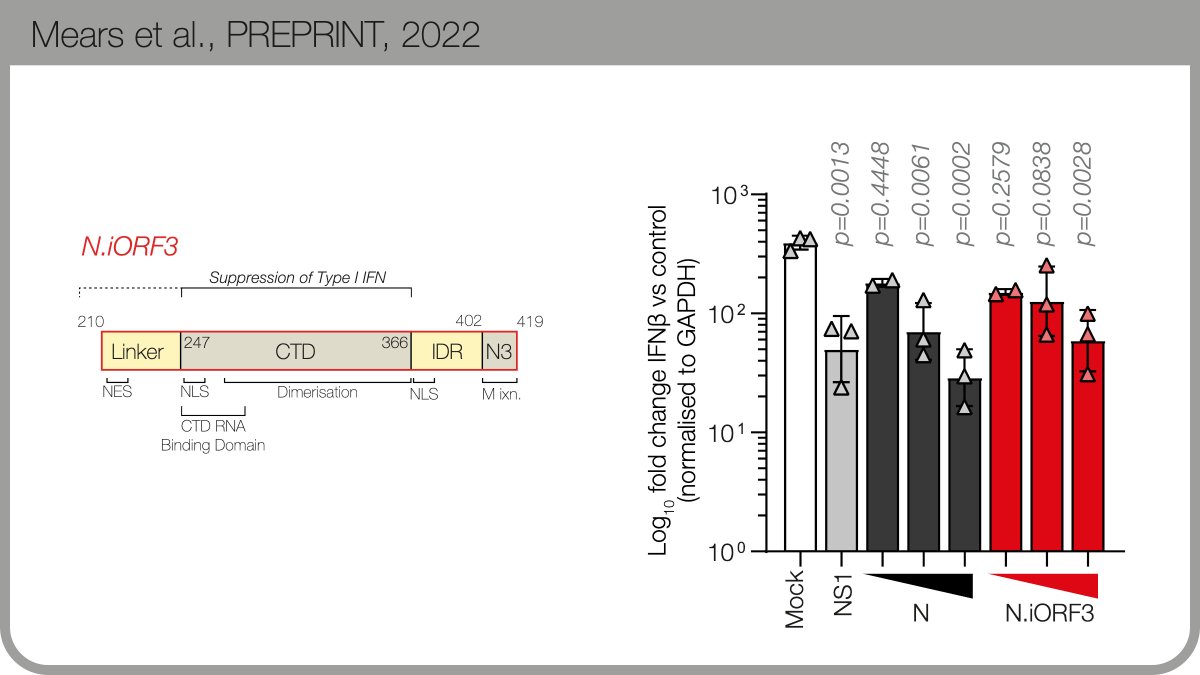
The protein product encodes by the new B.1.1.x sgmRNA is expressed during infection. We named it N.iORF3 following @SGinossarLab’s convention in nature.com. N.iORF3 encodes the C-terminal portion of nucleocapsid, which alone can suppress Type I interferon.
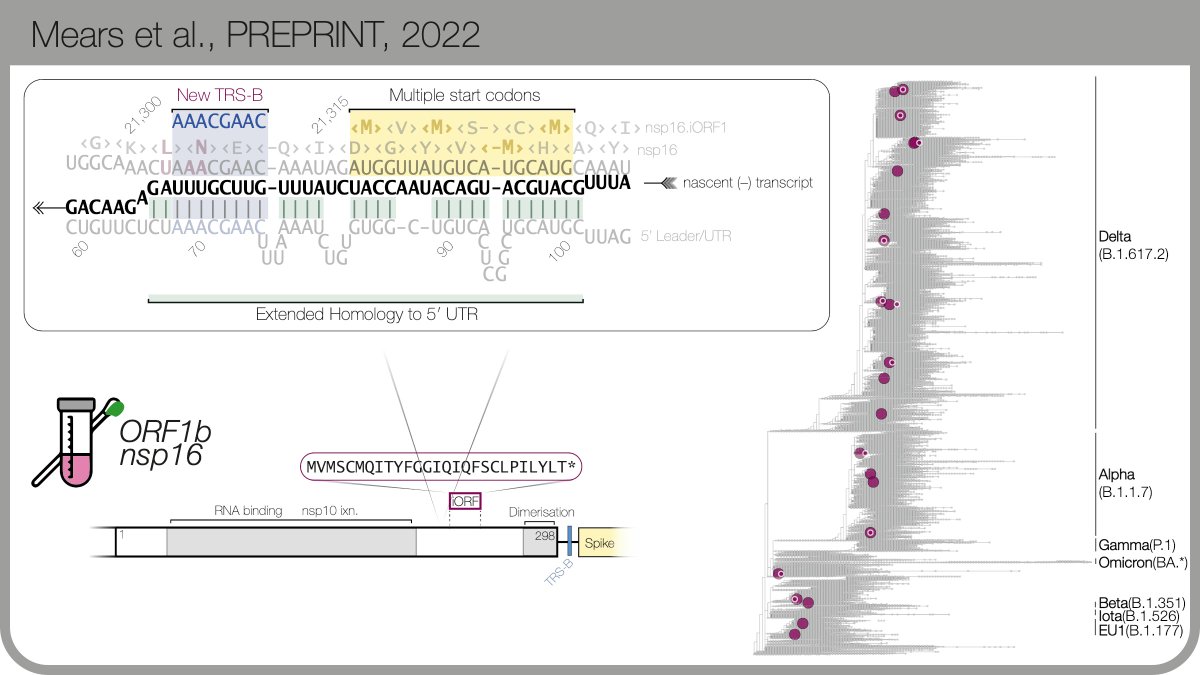
Thanks to @TheoSanderson and taxonium.org, we found that N.iORF3 evolved independently within a sublineage of the Iota variant. These aren’t artefacts: seqs cluster in geographic regions, show ongoing transmission & were detected by multiple depositing labs. 6/n
Given a new sgmRNA evolved >=2x, we wondered if another new sgmRNA had happened elsewhere unnoticed. 🤔 These changes aren’t annotated in @GISAID and there isn’t a “search for sequence” function, so #GeorgeYoung wrote one… 7/n
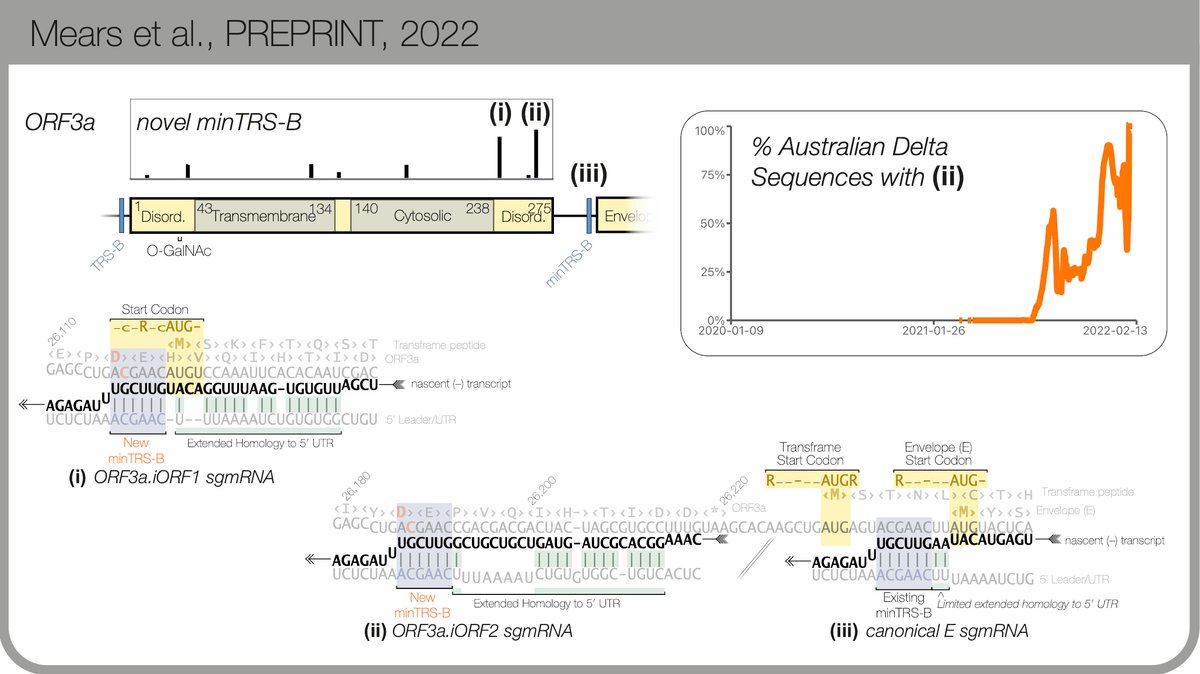
There are more examples (read the preprint! biorxiv.org) My favourite so far is within ORF3a, just upstream of E... before Omicron arrived, it had taken over all Delta sequences in 🇦🇺. 9/n
Together, our results suggests TRS site emergence within SARS-CoV-2 happens frequently on a global scale can generally lead to novel sgmRNA expression.
As I said before, this is “a thing” for coronaviruses... we just don’t usually get to see it “live”🧐. There’s also lots we don’t know about the function of these new sgmRNAs. @TheMenacheryLab and @doudna_lab have suggested a role for the amino acids in N.iORF3 TRS-B. 10/n
and Simon Mallal + @Thushan_deSilva + colleagues noticed the N.iORF3 sgmRNA in @CovidGenomicsUK data early on biorxiv.org ...
Also consequences for understanding past & future of the pandemic: 💡convergent evolution of TRSes makes it more challenging to trace the early evolution of SARS- CoV-2 in humans in which the N:203K,204R mutation was often used as a @PangoNetwork lineage-defining mutation. 13/n
Lastly, a thank you again to @Artisplicer & all who worked on this @TheCrick, incl @BABSBioinformat, ASF... and importantly @dremmacbw at the Legacy study, set up by @CharlesSwanton @SwantonLab @LabGandhi & Bryan Williams @UCLHresearch...
... who had the foresight to ensure that the "Legacy" of
@TheCrick's COVID-19 response extended to fundamental science. #MoreThanALifeboatLab. I am in awe of it all: hypothesis, to culture, and then to an archive of 750k clinical swabs. youtube.com. Thank you! 16/16
@TheCrick's COVID-19 response extended to fundamental science. #MoreThanALifeboatLab. I am in awe of it all: hypothesis, to culture, and then to an archive of 750k clinical swabs. youtube.com. Thank you! 16/16
Loading suggestions...