Technology
Health
Science
Machine Learning
Biotechnology
Protein structure prediction
Computational Biology
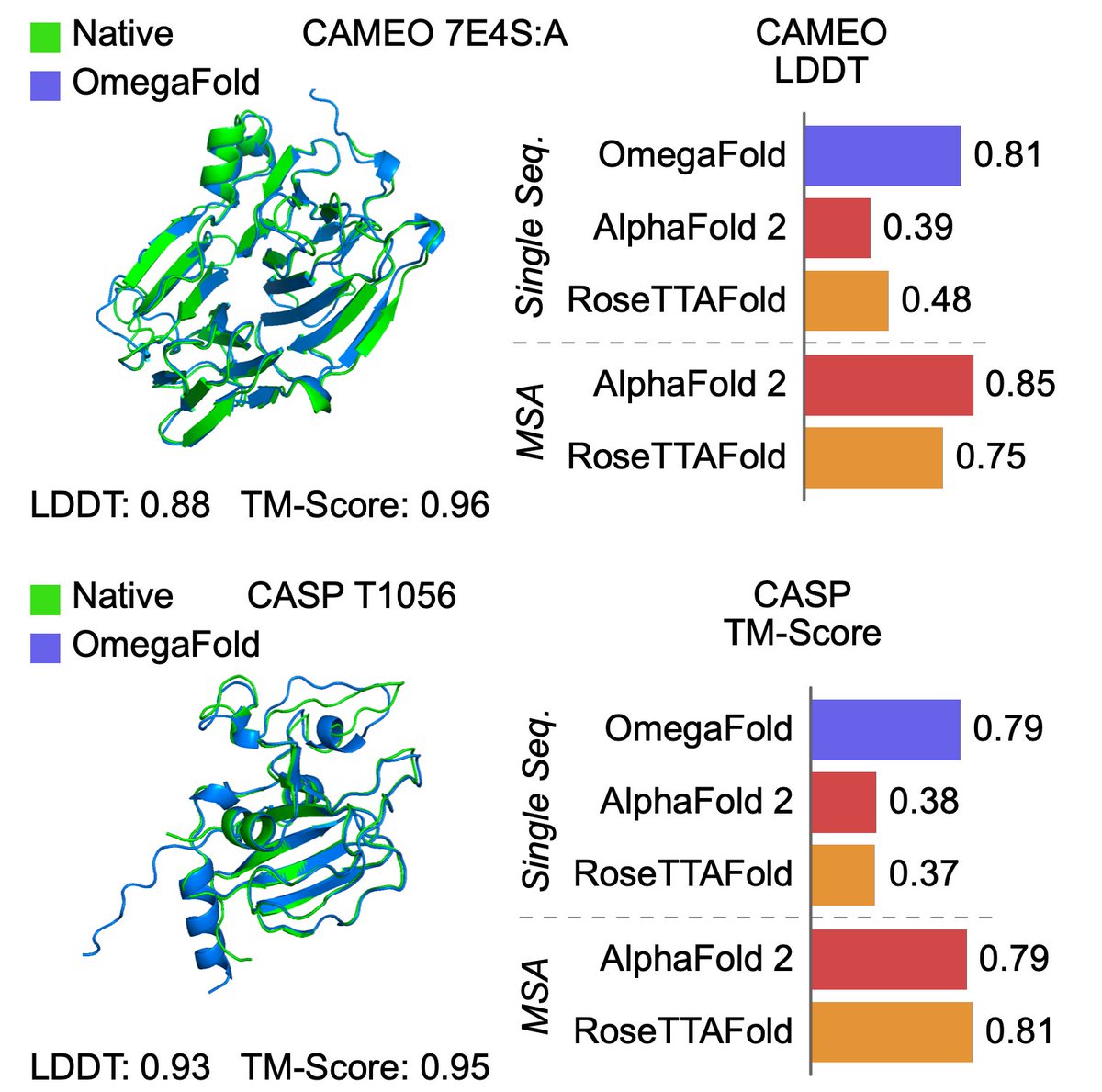
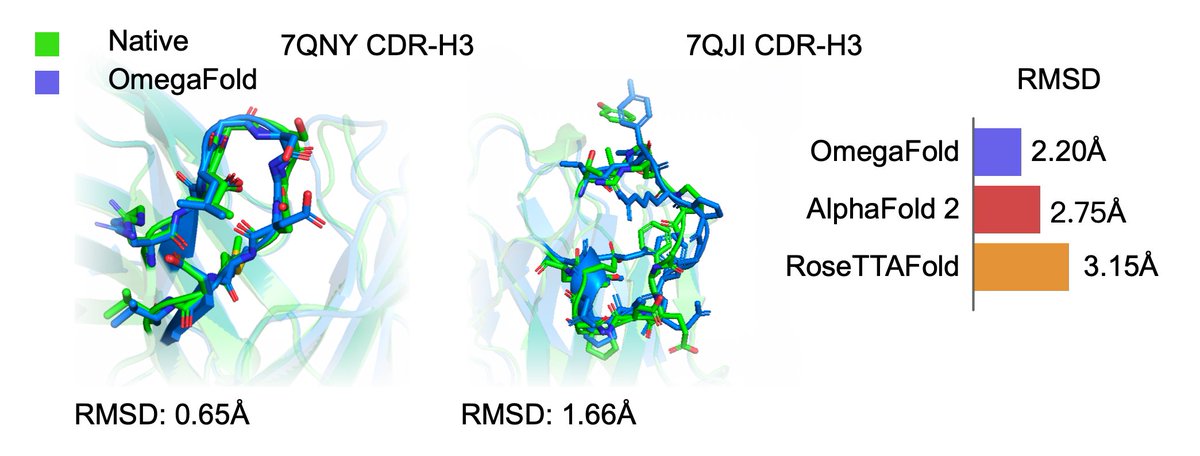
Protein structure can be predicted from a single sequence alone with high accuracy.
@HelixonBio team have developed OmegaFold, achieving performance similar to RF and AF2's MSA versions. Only a single sequence is given as input. 1/5
@HelixonBio team have developed OmegaFold, achieving performance similar to RF and AF2's MSA versions. Only a single sequence is given as input. 1/5
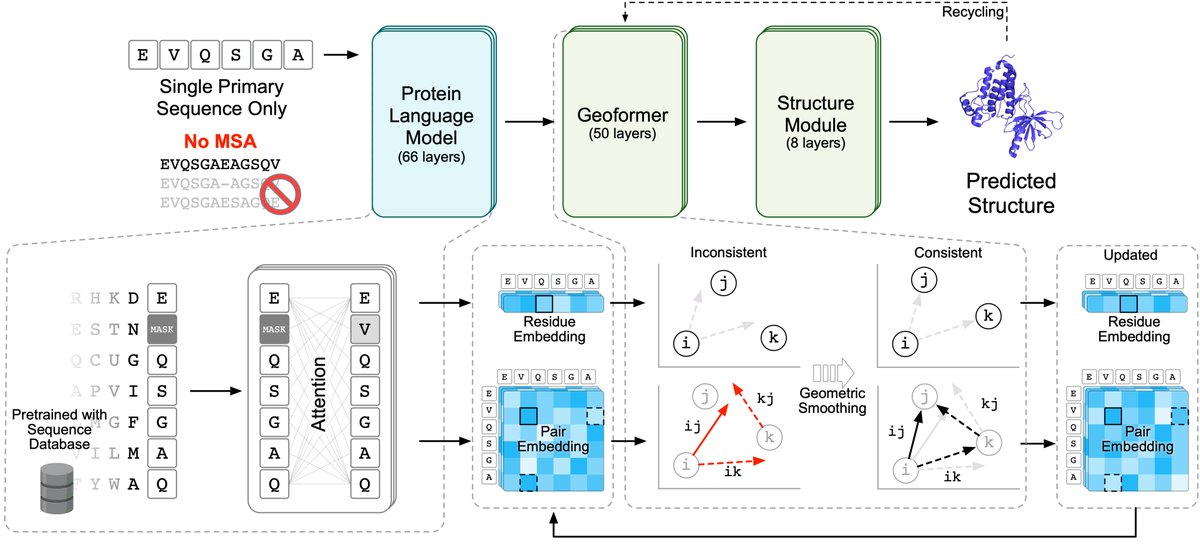
Protein language model (PLM) first intrinsically learns evolutionary context from large unaligned datasets. With new training techniques, we found improved long-range contact prediction. 3/5
Without the need to search MSAs, OF is also super fast. Only a few seconds for predicting a normal-size protein.
Preprint & code will be released soon. Just the pilot episode. Stay tuned for more. 5/5
Preprint & code will be released soon. Just the pilot episode. Stay tuned for more. 5/5
Loading suggestions...