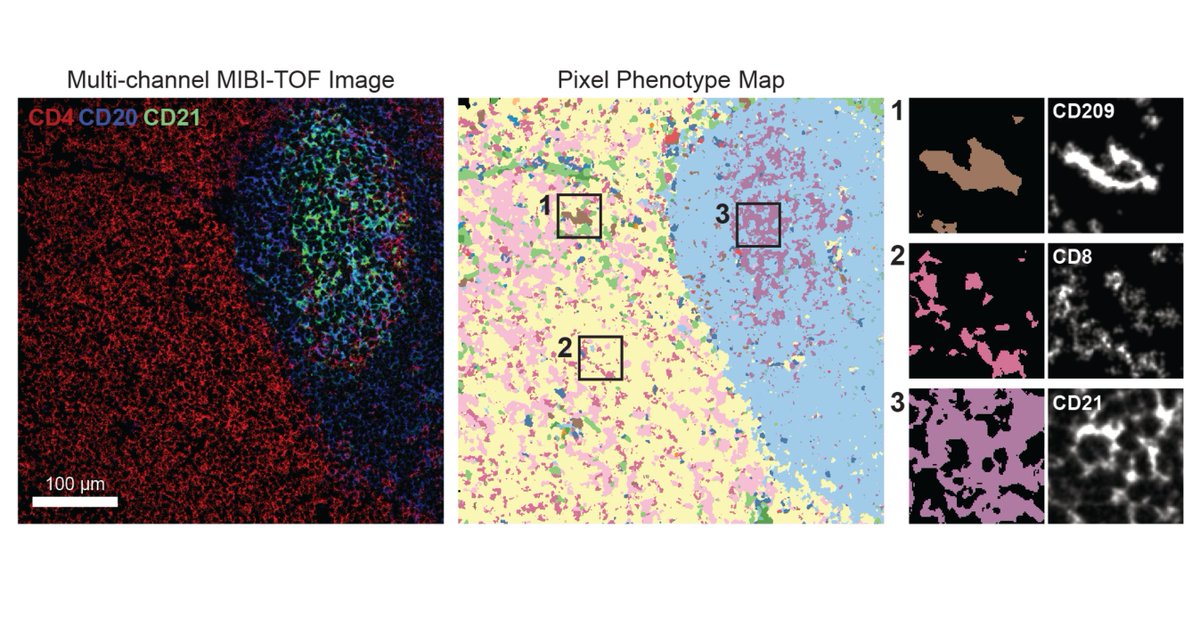
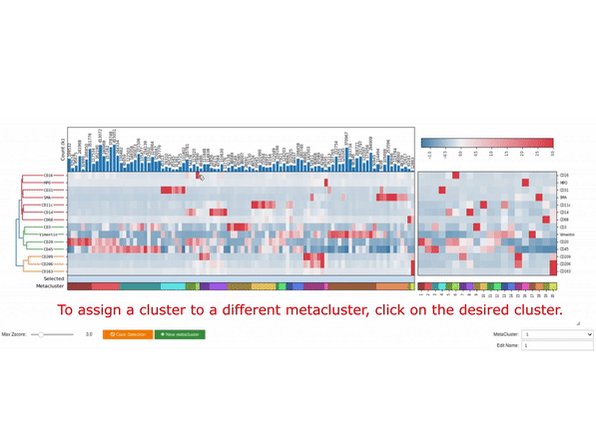
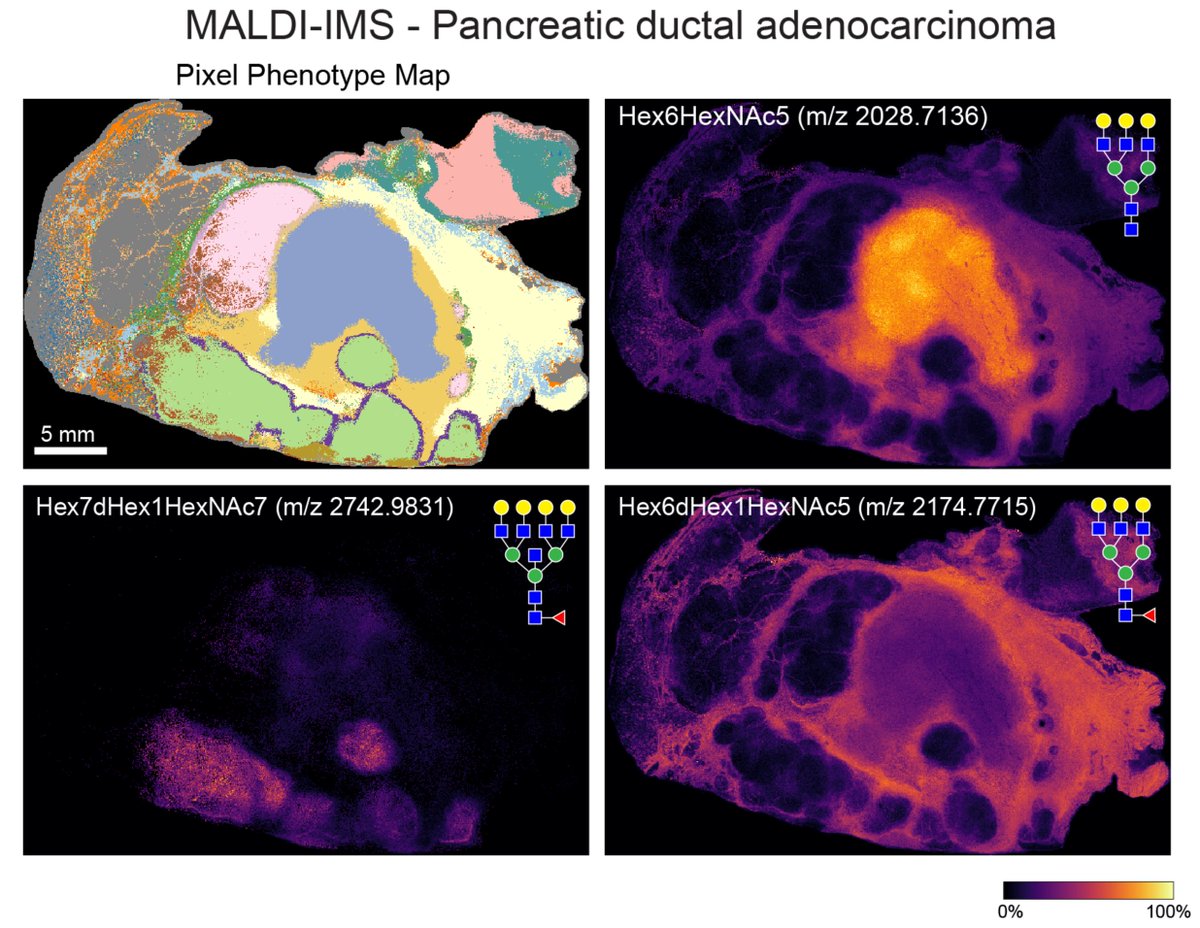
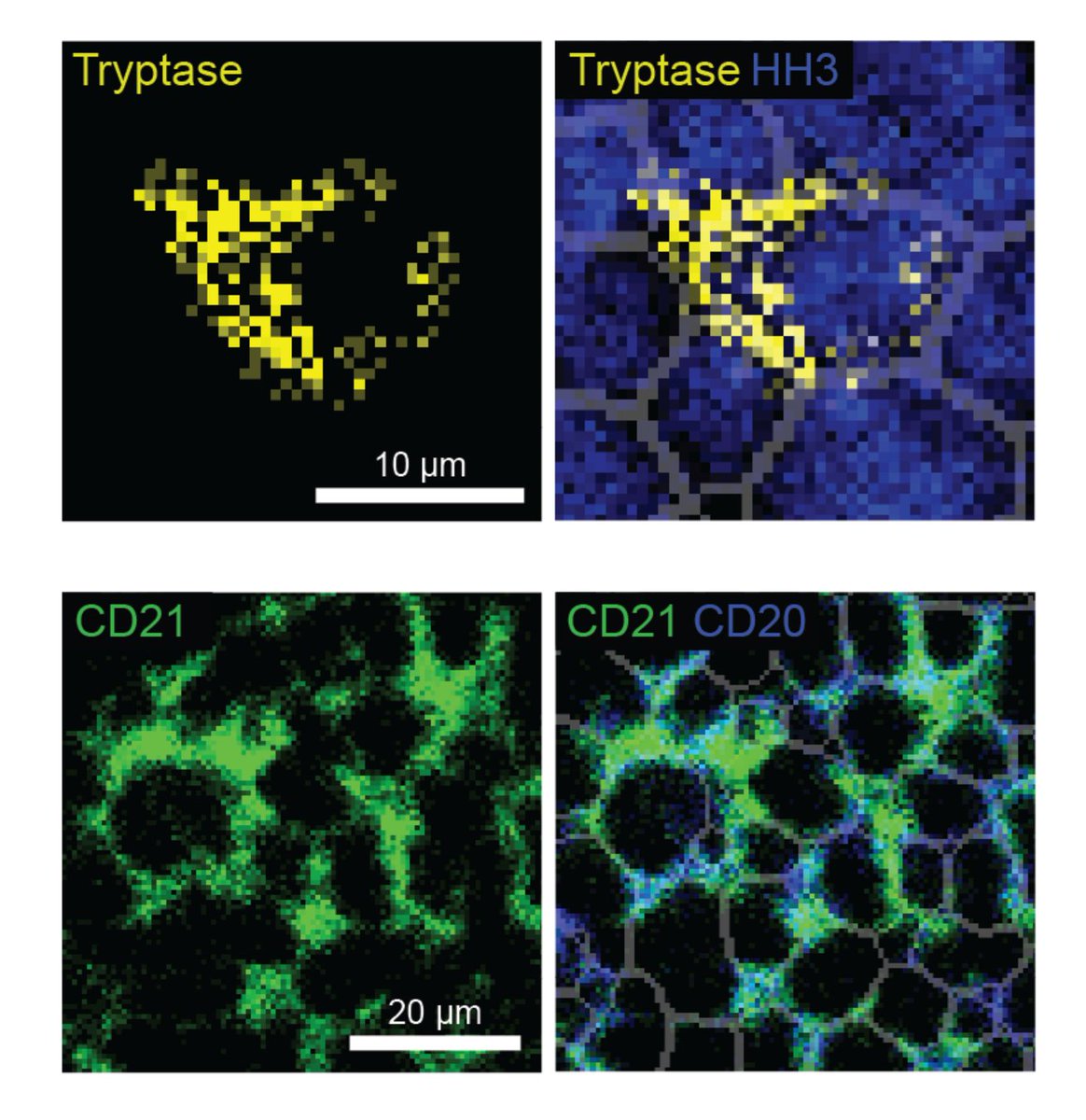
Excited to share the latest preprint with @MikeAngeloLab! While imaging studies often focus on cell objects, images capture significant info outside of cells. Enter Pixie, a pipeline for the quantitative annotation of both pixel and cell level features🧵biorxiv.org
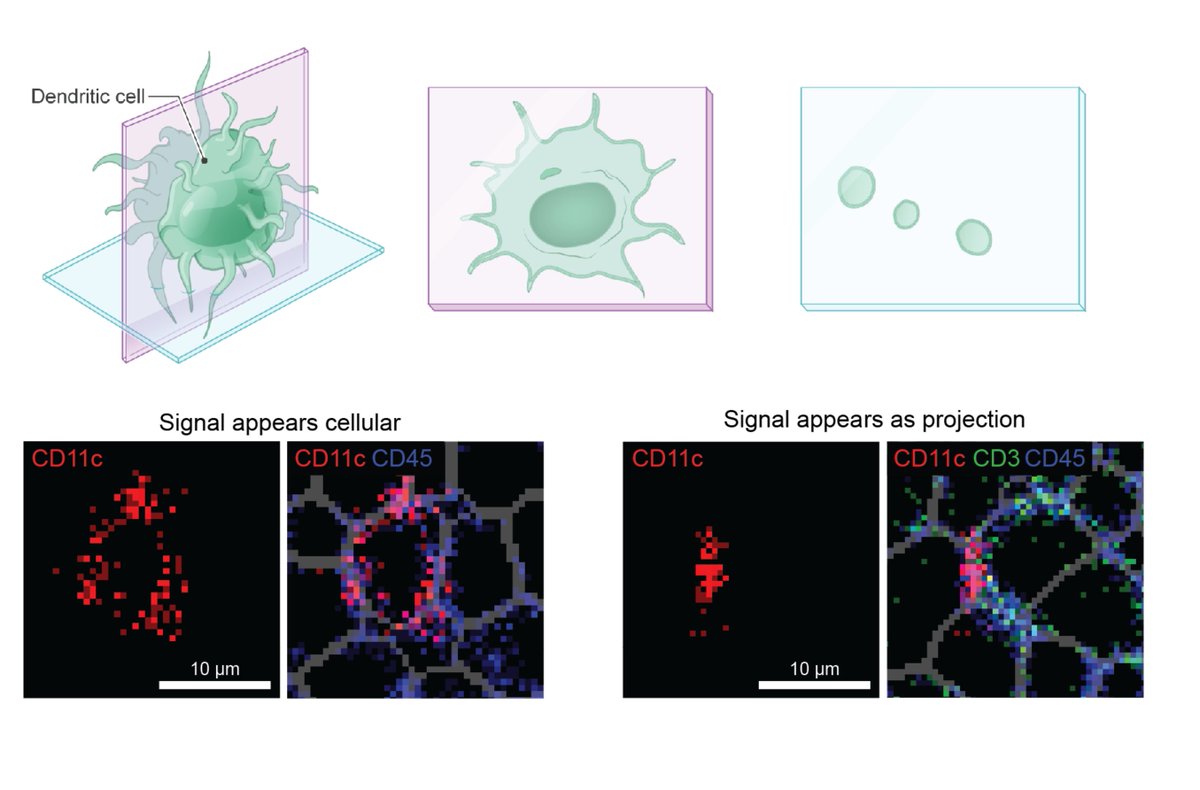
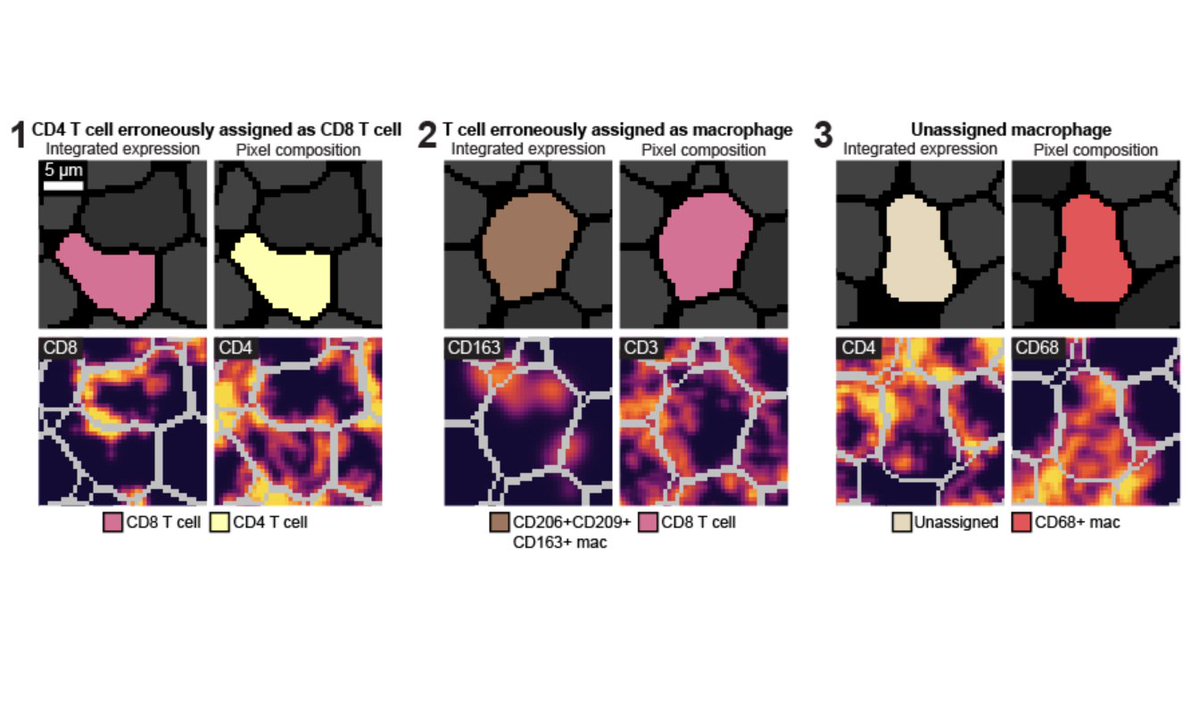
While segmentation algorithms (check out @NoahGreenwald's awesome work) can accurately identify cell boundaries in images, identifying cell phenotypes remains difficult, likely due to confounders such as plane of tissue sectioning, dense tissue, or irregularly shaped cells. [2/x]
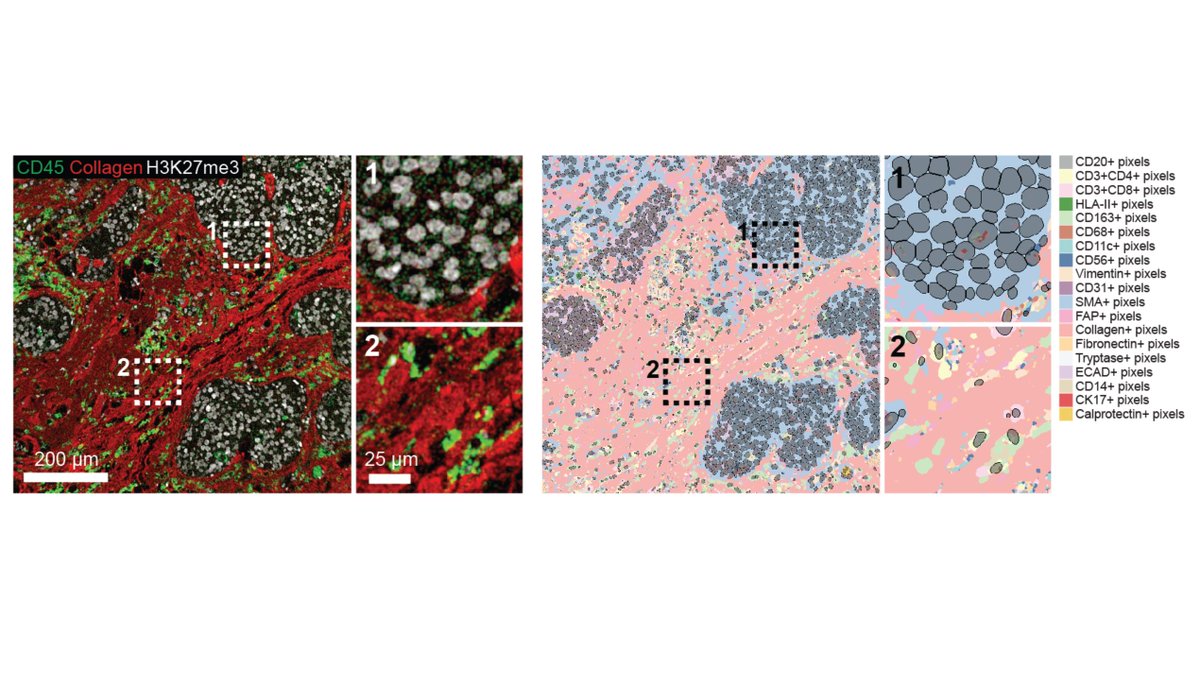
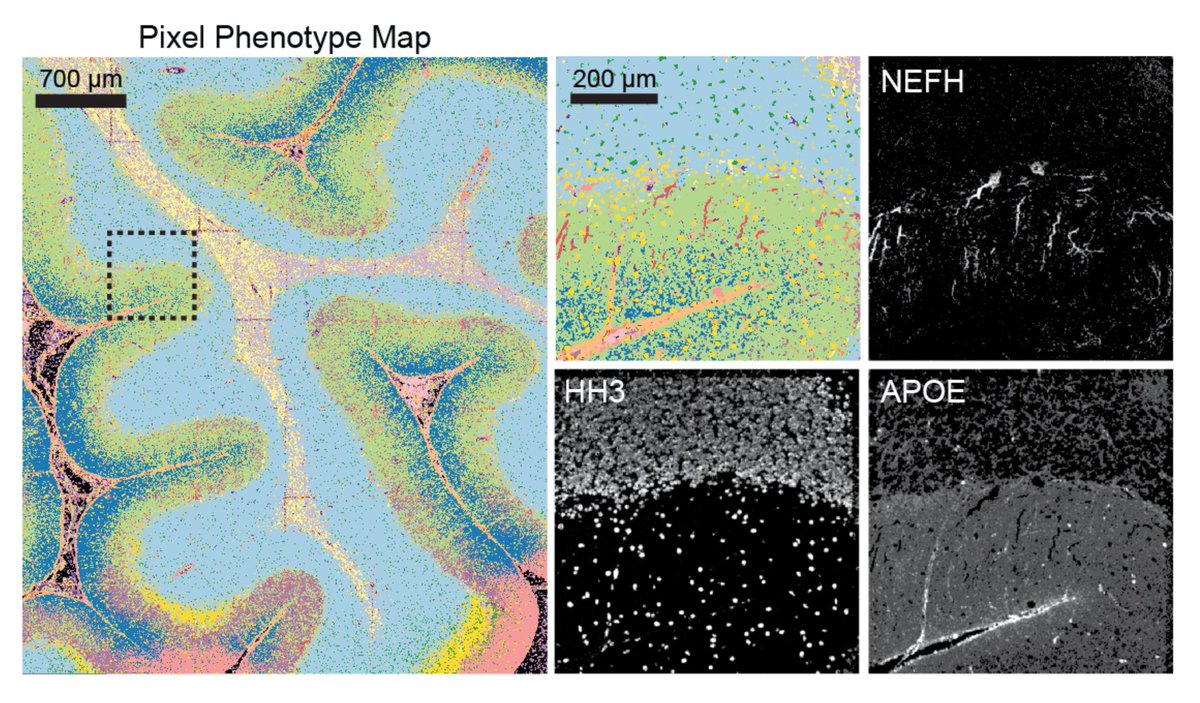
Due to abnormal shapes and complex spatial conformations of neuronal objects, cell segmentation is difficult in brain images. In beautiful MIBI images by @Dunja_Mrdjen, Pixie was able to map the full neuronal landscape, including neurons, vessels, astrocytes, and microglia. [7/x]
Taken together, Pixie is a simple, scalable pipeline that can generate quantitative annotations of features both independently and in conjunction with segmentation. Pixie is available as user-friendly Jupyter notebooks at github.com. Try it out! [11/x]
Big thanks to all my co-authors, @NoahGreenwald, Alex Kong, @ErinMcCaffrey14, Ke Leow, and @Dunja_Mrdjen. Who knew a quick "Can you try this one quick analysis for me?" from @MikeAngeloLab would turn into a whole paper? [12/end]
Loading suggestions...