Starting today, going to write threads on papers on non-genetic heritability & clonal heterogeneity (new + exciting areas) for a week.
Starting w/ @arjunrajlab ’s MemorySeq paper from 2020, which I think is crucial and quite a nice intro to the field:
cell.com
Starting w/ @arjunrajlab ’s MemorySeq paper from 2020, which I think is crucial and quite a nice intro to the field:
cell.com
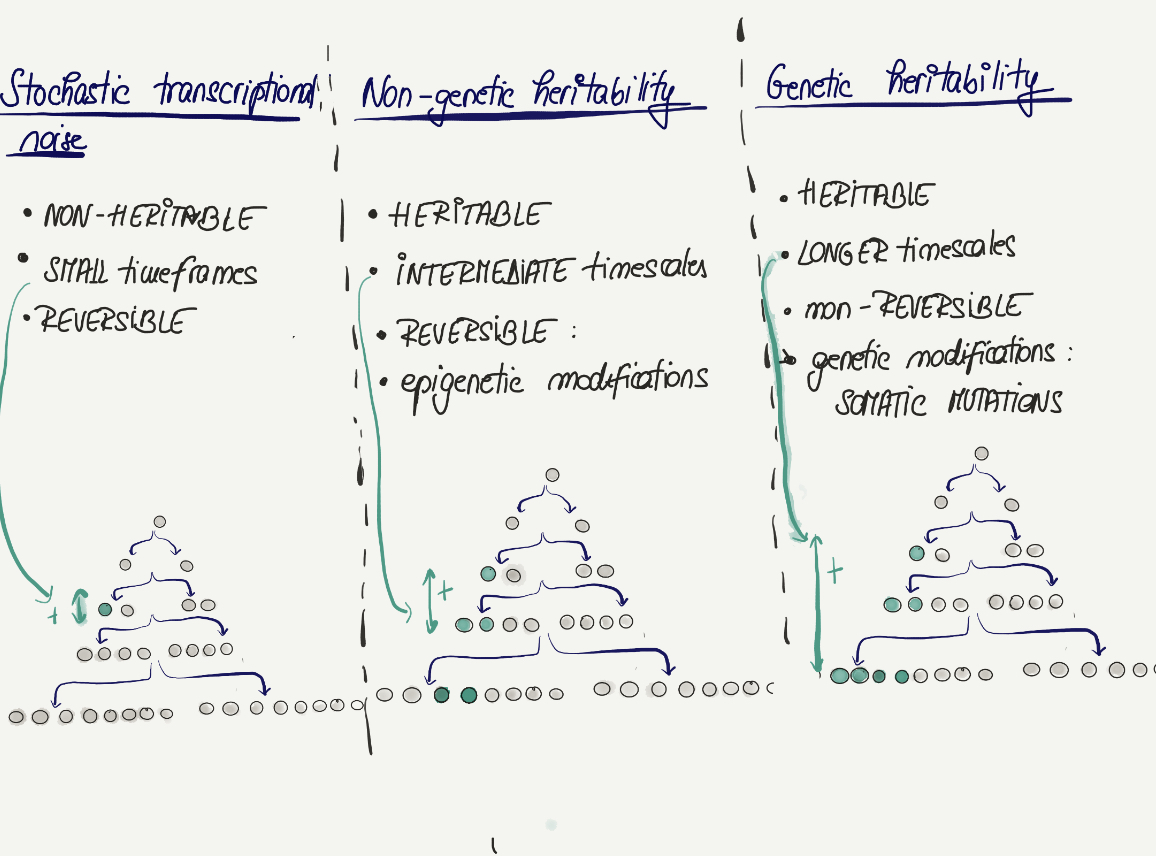
So what is non genetic heritability?
Cells can retain “memories” of gene expression according to the lineage they derive from & these memories are clonally transmitted.
One of the most obvious mechanisms of transmitting cellular memories is through permanent somatic mutations
Cells can retain “memories” of gene expression according to the lineage they derive from & these memories are clonally transmitted.
One of the most obvious mechanisms of transmitting cellular memories is through permanent somatic mutations
This has been widely studied, particularly in the context of cancer, where somatic mutations lead to clonal expansions and tumour progression.
Non generic heritability aims to quantify a different type of cellular “memory”: one that lies on an intermediate scale
Non generic heritability aims to quantify a different type of cellular “memory”: one that lies on an intermediate scale
How does MemorySeq aim to quantify such intermediate timescale cellular memory?
Single cells from a melanoma cell line (WM989-A6) were seeded into 48 separate wells and allowed to proliferate. The clones were then subjected to RNASeq.
Single cells from a melanoma cell line (WM989-A6) were seeded into 48 separate wells and allowed to proliferate. The clones were then subjected to RNASeq.
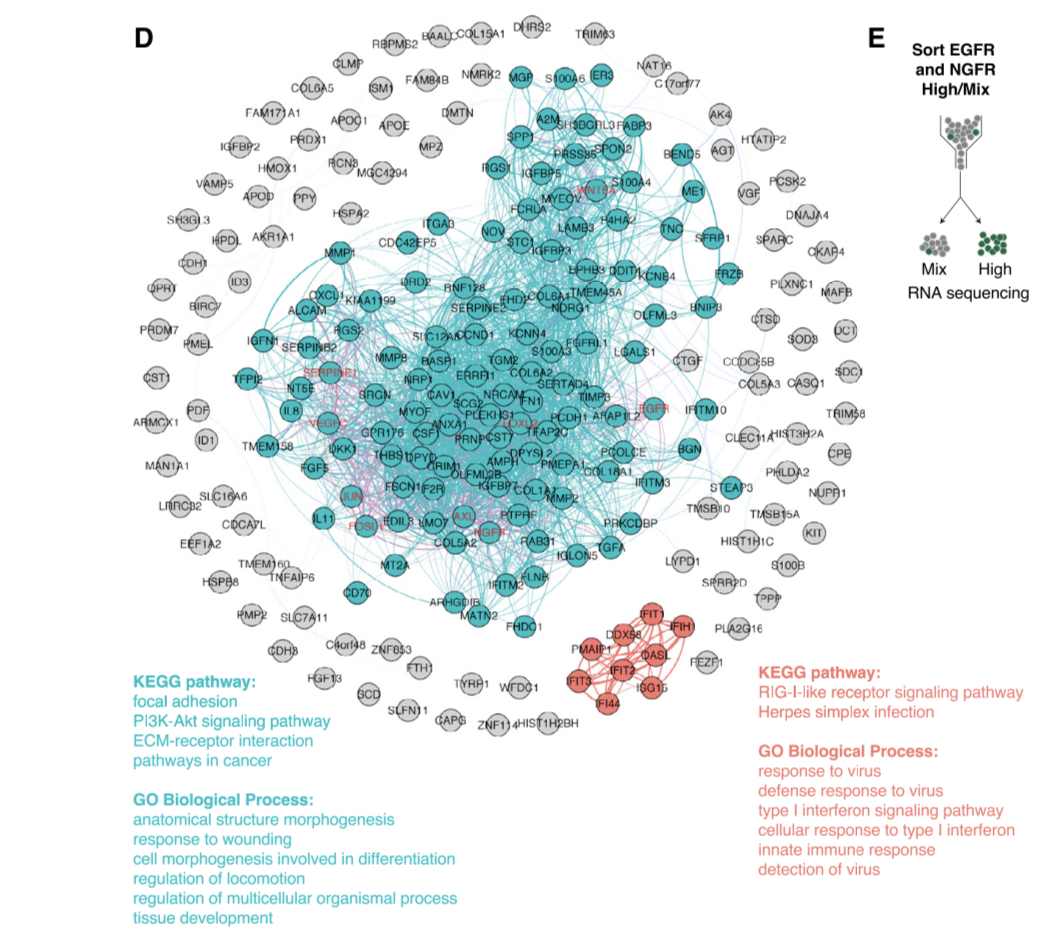
Crucially, genes which had previously been associated with resistance to cancer in this specific melanoma type (EGFR, NGF) showed higher levels of variability. By contrast housekeeping genes showed low variability.
This hints at a possible biological role of this heterogeneity.
This hints at a possible biological role of this heterogeneity.
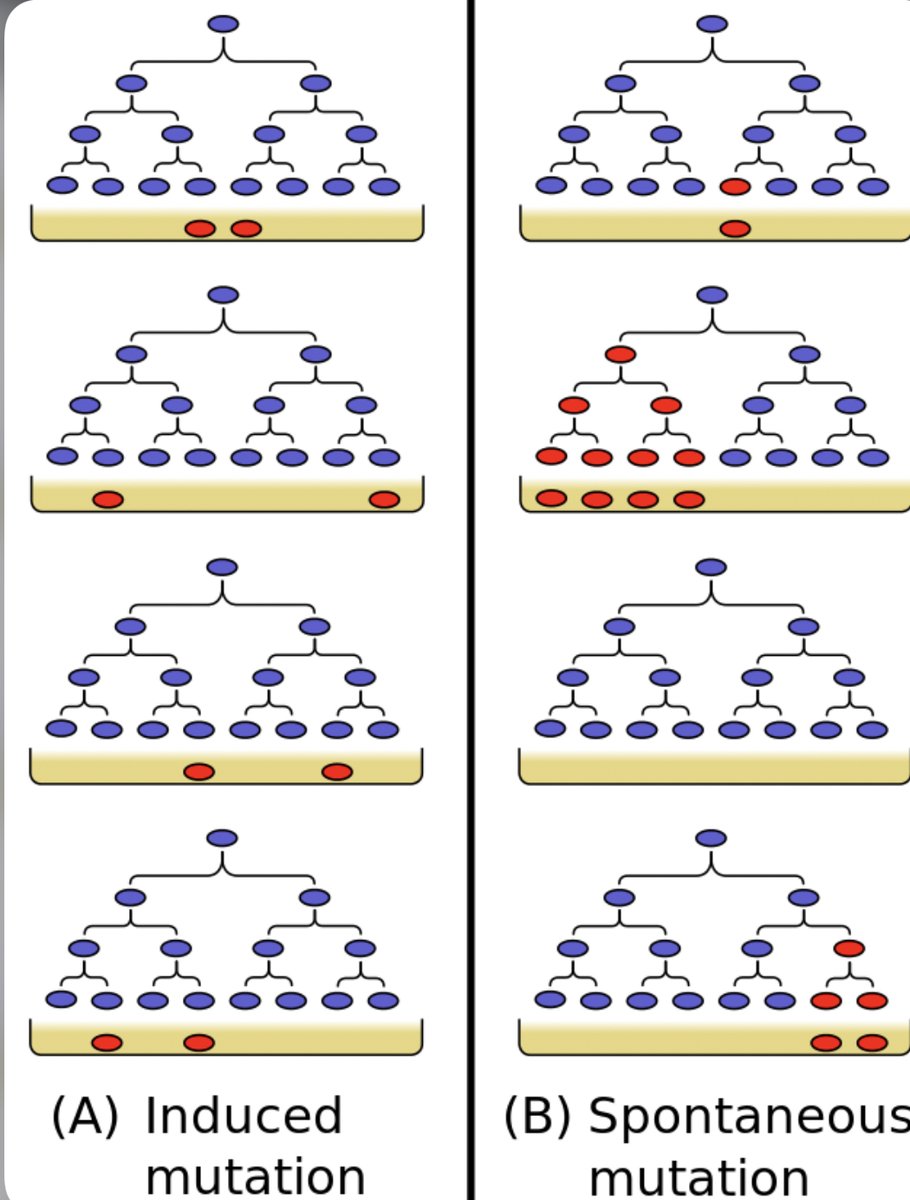
Then, the authors tried to directly confirm the key prediction of MemorySeq:
That a gene which shows high variability across MemorySeq clones would occasionally initiate high levels of expression that can persist across multiple cell divisions but don’t persist indefinitely.
That a gene which shows high variability across MemorySeq clones would occasionally initiate high levels of expression that can persist across multiple cell divisions but don’t persist indefinitely.
The final part of the paper addresses what might be the most interesting question posed by this paper: what might be the biological implications of such non- genetic heritability?
As hinted by the types of genes that are “affected” by this, a candidate could be drug resistance
As hinted by the types of genes that are “affected” by this, a candidate could be drug resistance
This strongly implicates Non genetic heritability as having a role in drug resistance.
Heritable expression of certain genes would lead to functionally distinct subpopulations that could account for drug resistance.
Thus, MemorySeq could serve as a tool of IDing these subpopulations that are not easy to identify via other methods.
Thus, MemorySeq could serve as a tool of IDing these subpopulations that are not easy to identify via other methods.
An interesting obs. :
Heritable gene expression changes could be more powerful biologically if instead of affecting only one gene at once, they would act on multiple genes.
The impacts of these co-variables genes would thus re-enforce each other to produce a greater effect.
Heritable gene expression changes could be more powerful biologically if instead of affecting only one gene at once, they would act on multiple genes.
The impacts of these co-variables genes would thus re-enforce each other to produce a greater effect.
So do such co-fluctuating gene modules (which make sense in theory) exist?
Notably, some of the genes within an individual “heritability module” are scattered across different chromosomes, raising an important question as to the mechanism that allows for such co-fluctuation.
Identifying the molecular mechanism is very briefly attempted in the paper, but it’s more of an afterthought (which is understandable given how complex the paper already is).
This is not to criticise the paper, but rather to highlight that identifying the mechanisms behind this non-genetic heritability is one of the major open questions posed by this paper.
The paper does briefly point towards some transcription factors and histone mods.
The paper does briefly point towards some transcription factors and histone mods.
Of 2 Events co-occurring.
However, it’s suggested the expression of genes organised in “modules” is correlated (so not independent events) & that there are biological mechanisms that underpin their co-expression across chromosomes.
However, it’s suggested the expression of genes organised in “modules” is correlated (so not independent events) & that there are biological mechanisms that underpin their co-expression across chromosomes.
Overall I liked this paper a lot & I found the initial MemorySeq experiment to be particularly elegant.
I think it’s a nice intro to the very new field of non-generic heritability.
I think it’s a nice intro to the very new field of non-generic heritability.
I hope to explore other papers in the coming days and show how non-genetic heritability is relevant to a bunch of questions beyond drug resistance which include: cell type classification, differentiation, immune response + somatic evolution.
Loading suggestions...