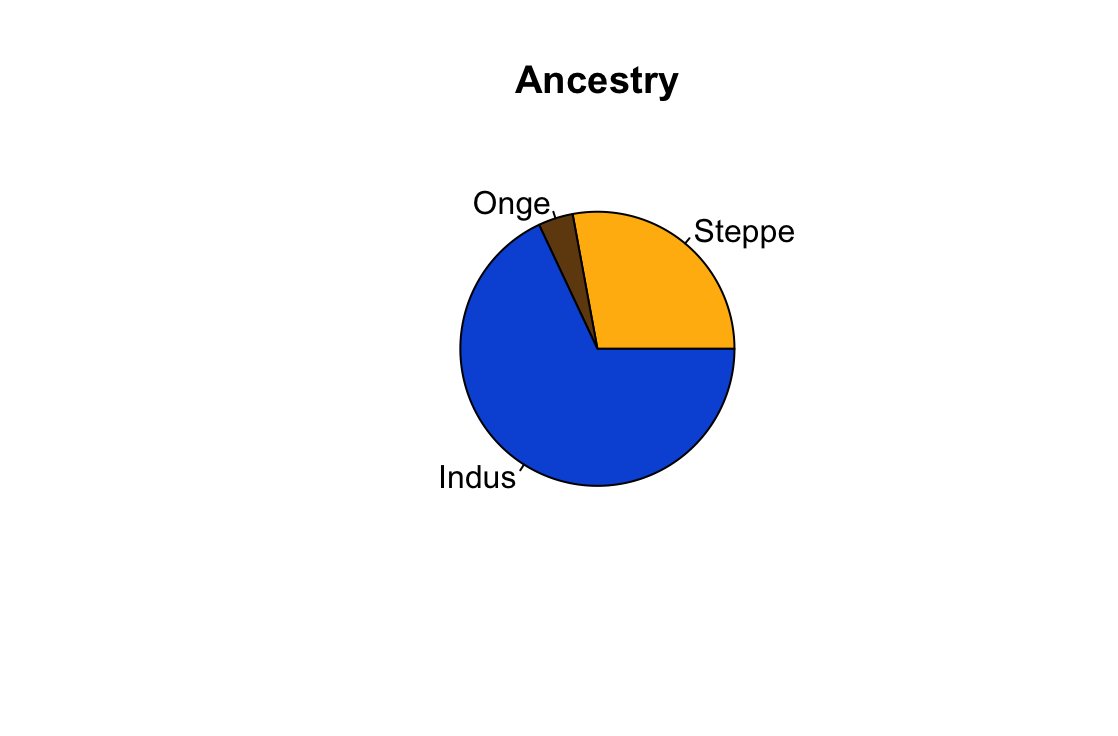
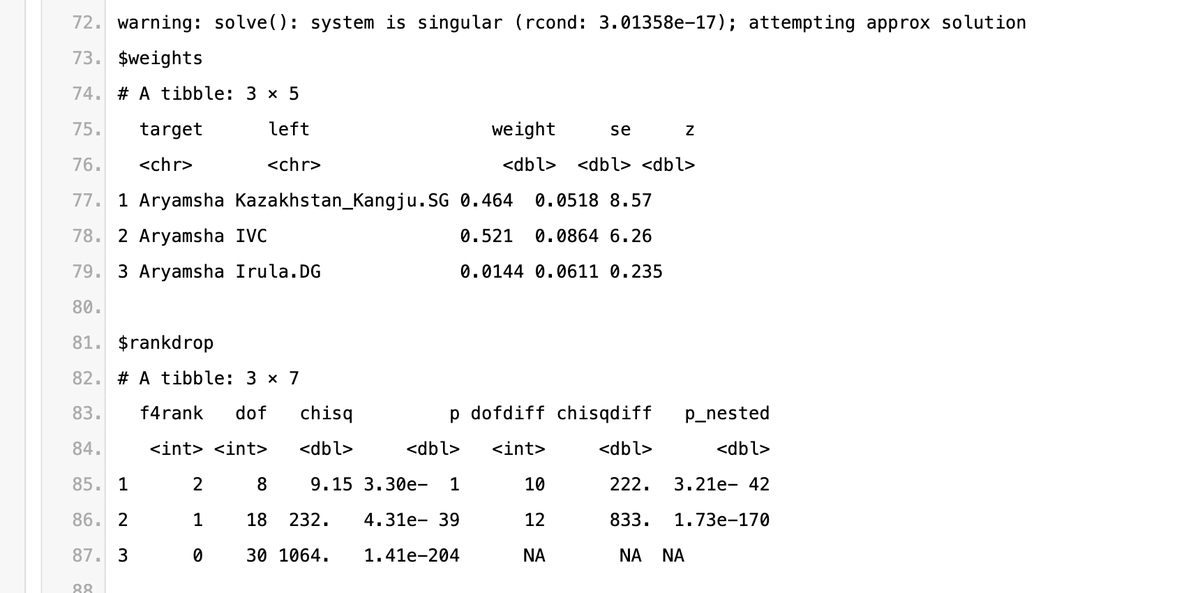
Here's a pastebin file containing all the qpadm runs. Unfortunately the coverage is 23andme V5 which is poor and gives just gives 133k snps but it is sufficient enough to see that G25 is kinda wonky in many things.
Few notes on this below..+
pastebin.com
Few notes on this below..+
pastebin.com
1) Kangju from historical era works as the best Steppe source, giving the highest p-values.
Other Steppe sources that work just about as well are Andronovo ones mixed with some Siberian like DashtyKozy, Kashikarchi and Taldysay MLBA.
Other Steppe sources that work just about as well are Andronovo ones mixed with some Siberian like DashtyKozy, Kashikarchi and Taldysay MLBA.
Loading suggestions...