🧵Intro to CRISPR
Here we will look at the discovery of CRISPR.
Here we will look at the discovery of CRISPR.
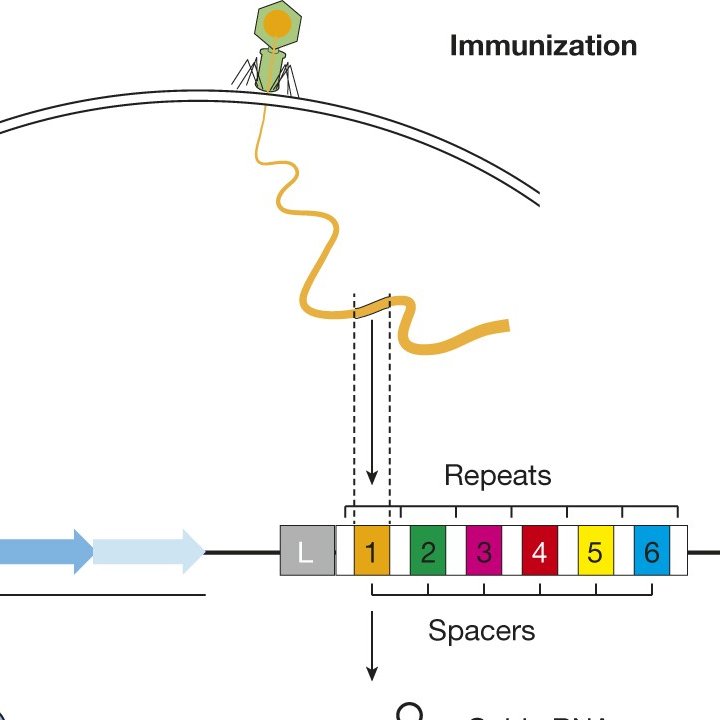
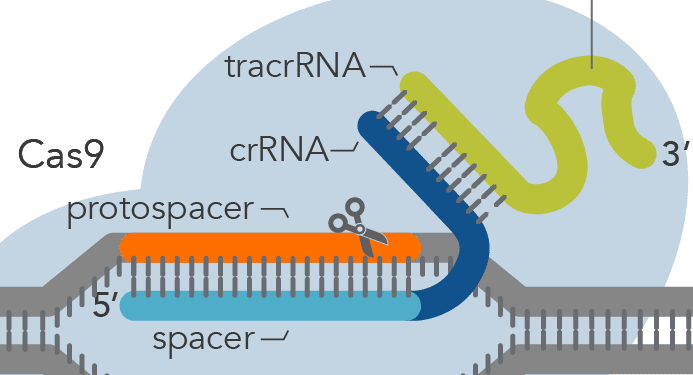
6/ The crRNA will then bind to the tracrRNA to make a complete guide RNA. The two RNA segments bind together by complementary base pair binding by hydrogen bonding as shown in the picture above.
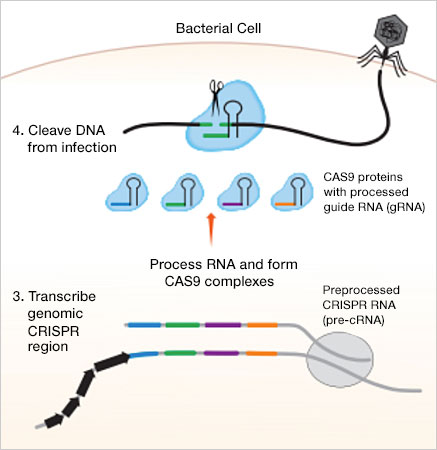
8/ The actual guide sequence is about 20 nucleotides long. Once it finds the matching viral DNA, the CAS9 enzyme will cut that DNA. This destroys the viral DNA.
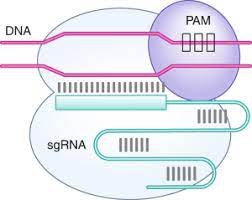
10/ The answer to that is the PAM sequence of the CAS9 enzyme. This stands for Protospacer Adjacent Motif. This is a small segment of nucleotides in the DNA that is recognized by the CAS9 enzyme which is N-G-G.
11/ The N stands for any nucleotide while the G stands for Guanine. This basically means the CAS9 enzyme not only needs to match the guide RNA sequence to the protospacer, but it also needs to match its PAM to the 2 Guanines near the protospacer.
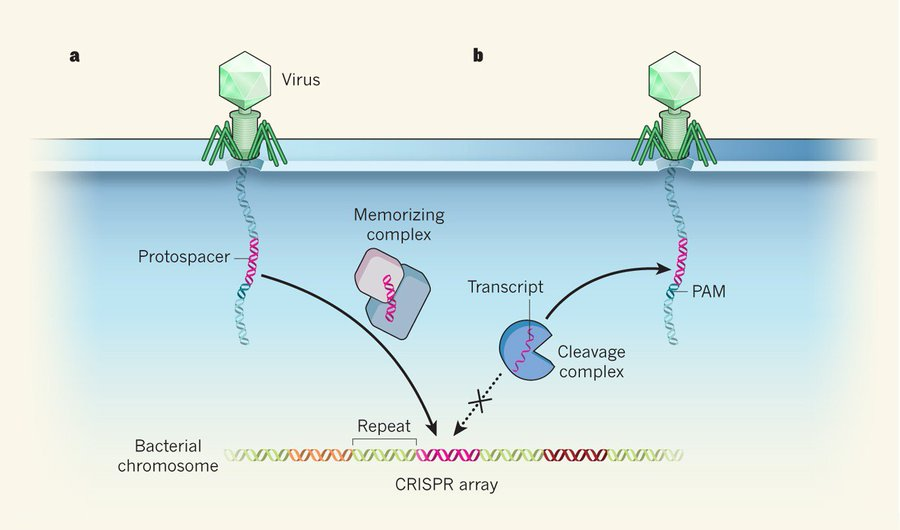
12/ If both don't exist, the CAS9 will not cut. As you can guess, the CRISPR region of the bacteria genome would not contain the PAM sequence so the CAS9 would be selective for targeting the viral DNA, but not the bacteria's DNA.
Loading suggestions...