🧬 Preprint 🧬
Most mutations begin as a change in only one of the two DNA strands.
To see these single-strand precursors of mutations, we developed single-molecule sequencing that achieves single-molecule and *single-strand* fidelity.
biorxiv.org
🧵⬇️
1/11
Most mutations begin as a change in only one of the two DNA strands.
To see these single-strand precursors of mutations, we developed single-molecule sequencing that achieves single-molecule and *single-strand* fidelity.
biorxiv.org
🧵⬇️
1/11
Led by superstar @meihliu, with fearless work by @benmcosta and Una Choi, and many others.
Grateful to our close collaborator @jeshoag and to @Uri_Tabori, @Cryos_Int, and @ResearchCM.
🧵 ⬇️
2/11
Grateful to our close collaborator @jeshoag and to @Uri_Tabori, @Cryos_Int, and @ResearchCM.
🧵 ⬇️
2/11
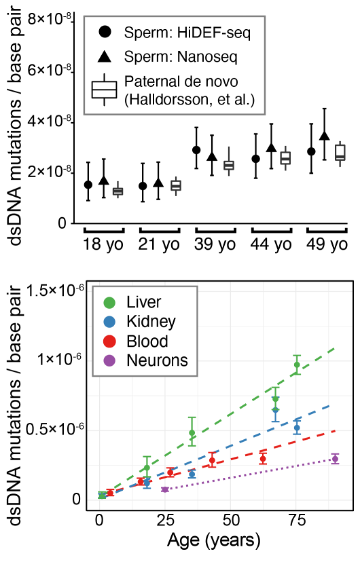
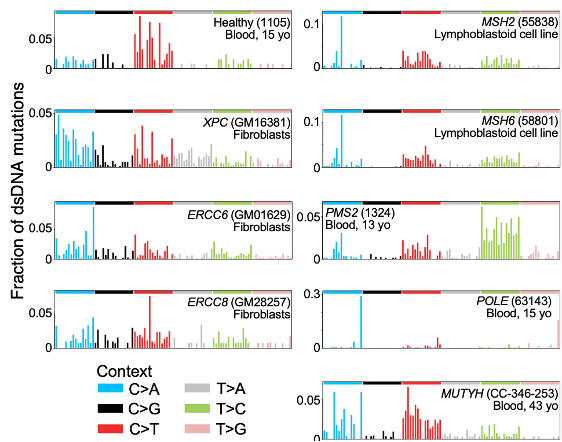
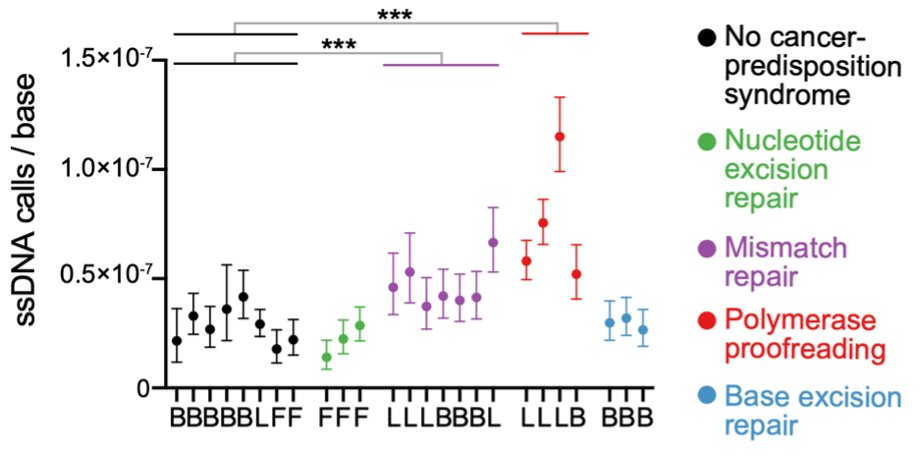
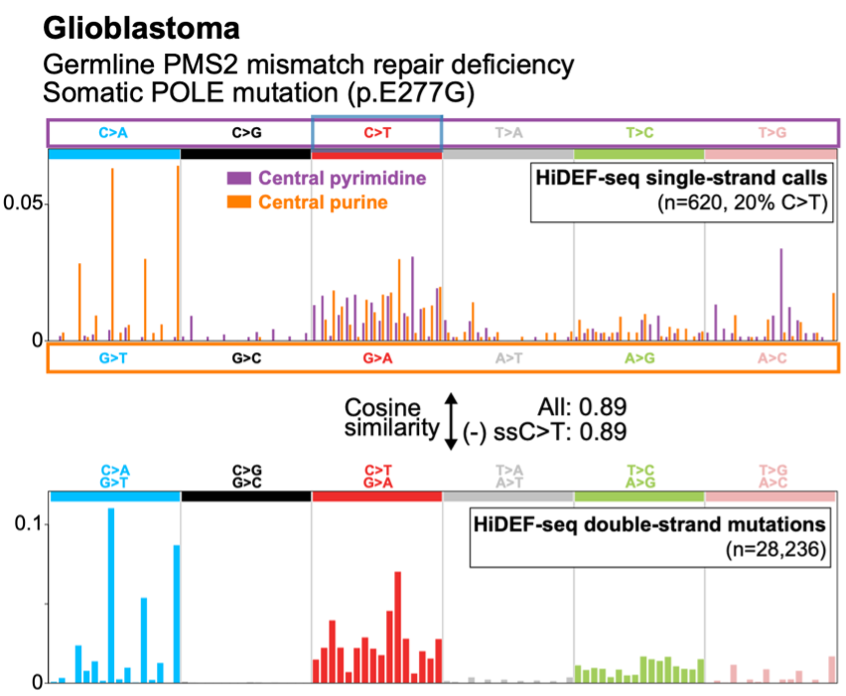
Mosaic mutations are challenging to detect, since each may be present in only one DNA molecule (in one cell). So we need single-molecule fidelity.
Duplex-seq methods like NanoSeq by Abascal, et al, and Single-cell DNA-seq methods achieve this.
3/11
Duplex-seq methods like NanoSeq by Abascal, et al, and Single-cell DNA-seq methods achieve this.
3/11
But current high-fidelity methods only reliably detect mutations present in *both* DNA strands.
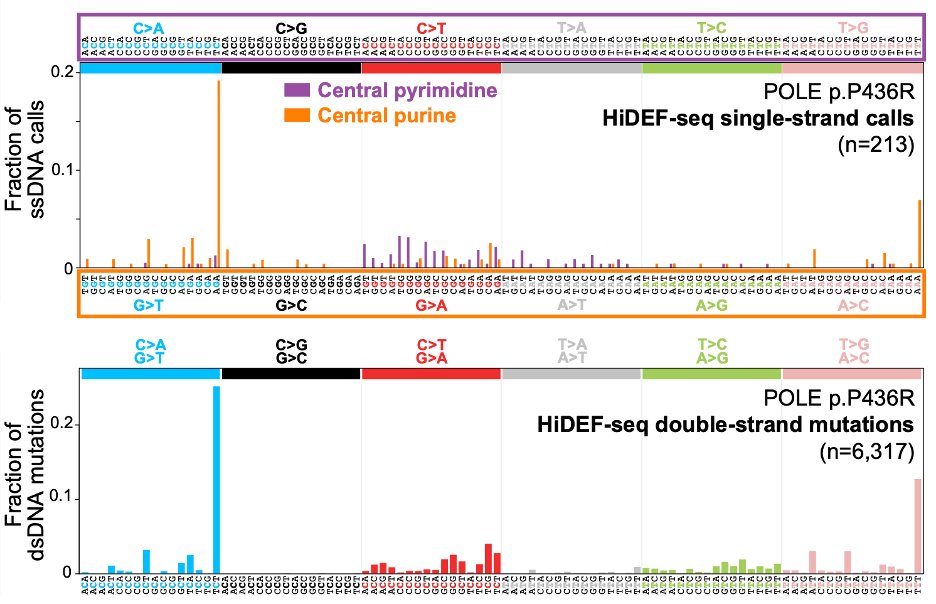
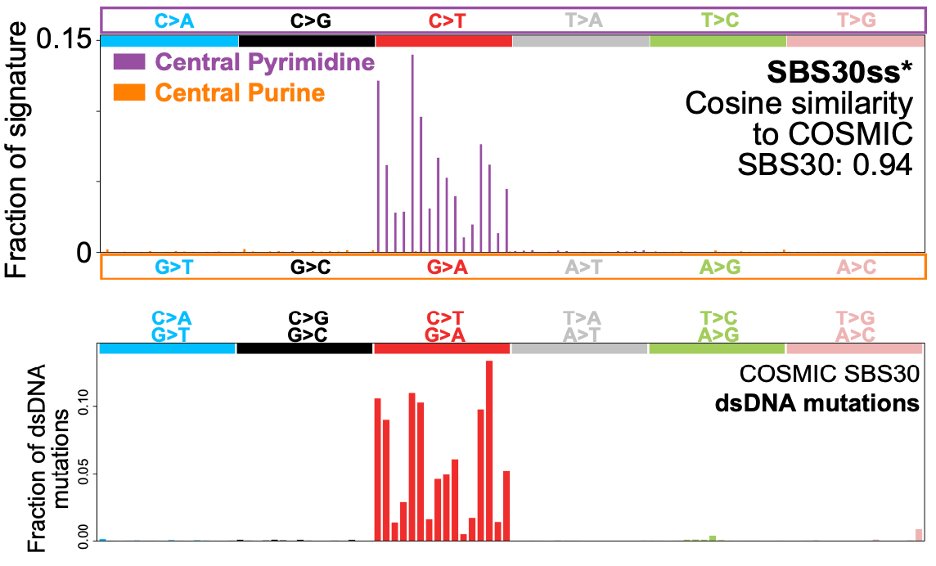
Our approach, which we call HiDEF-seq, achieves single-molecule accuracy for single-base substitutions in either one or both strands, by direct sequencing of single molecules.
4/11
Our approach, which we call HiDEF-seq, achieves single-molecule accuracy for single-base substitutions in either one or both strands, by direct sequencing of single molecules.
4/11
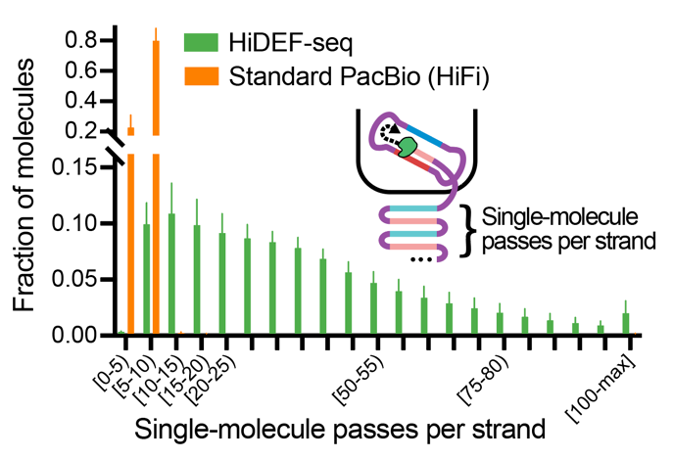
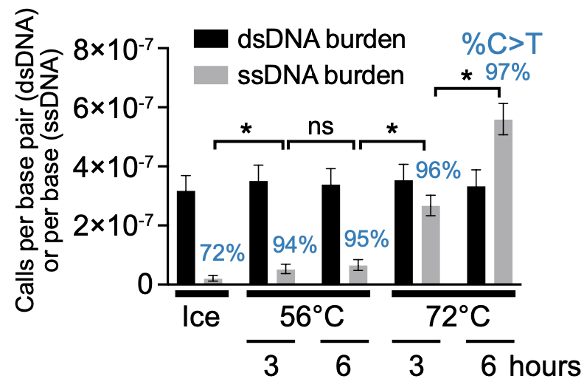
The recipe took a few years to hone: ⬆️ seq “passes” per molecule in @PacBio libraries, ligating nicks, blocking residual nicks (🙏 NanoSeq), a few other mol bio tricks, and a single-strand computational pipeline.
5/11
5/11
See the paper for more! We hope HiDEF-seq will be used in many new kinds of studies to understand the origins of mutations.
Deeply grateful to our funders @sontagfdn, @NIH, @pewtrusts.
11/11
Deeply grateful to our funders @sontagfdn, @NIH, @pewtrusts.
11/11
Loading suggestions...