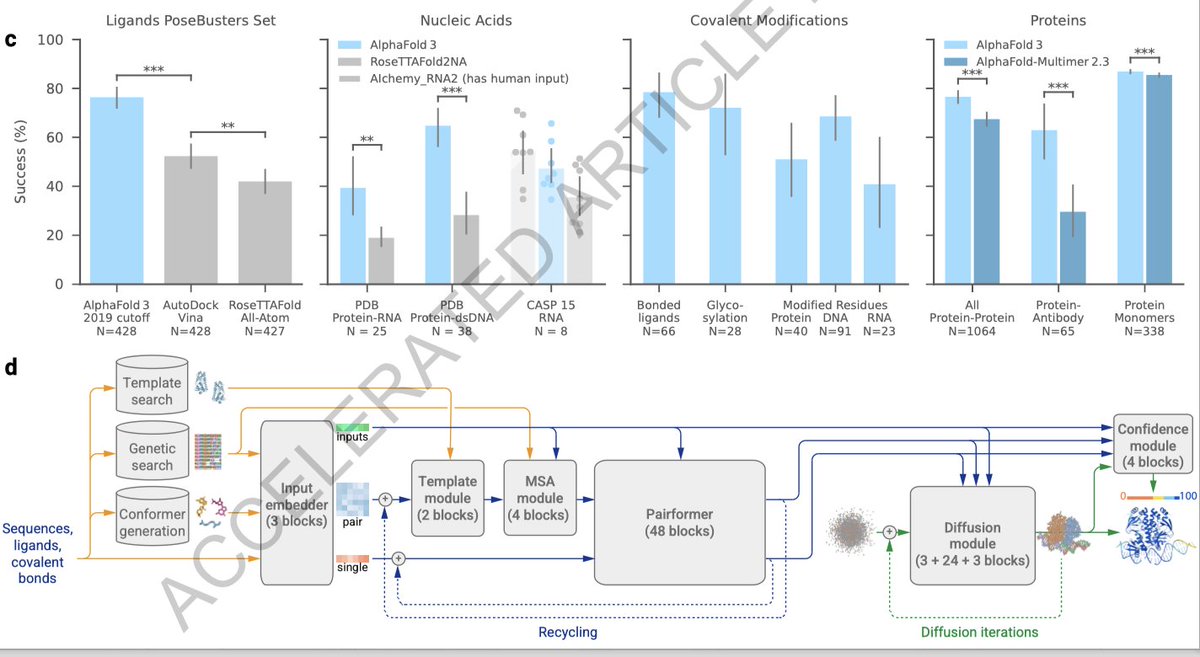
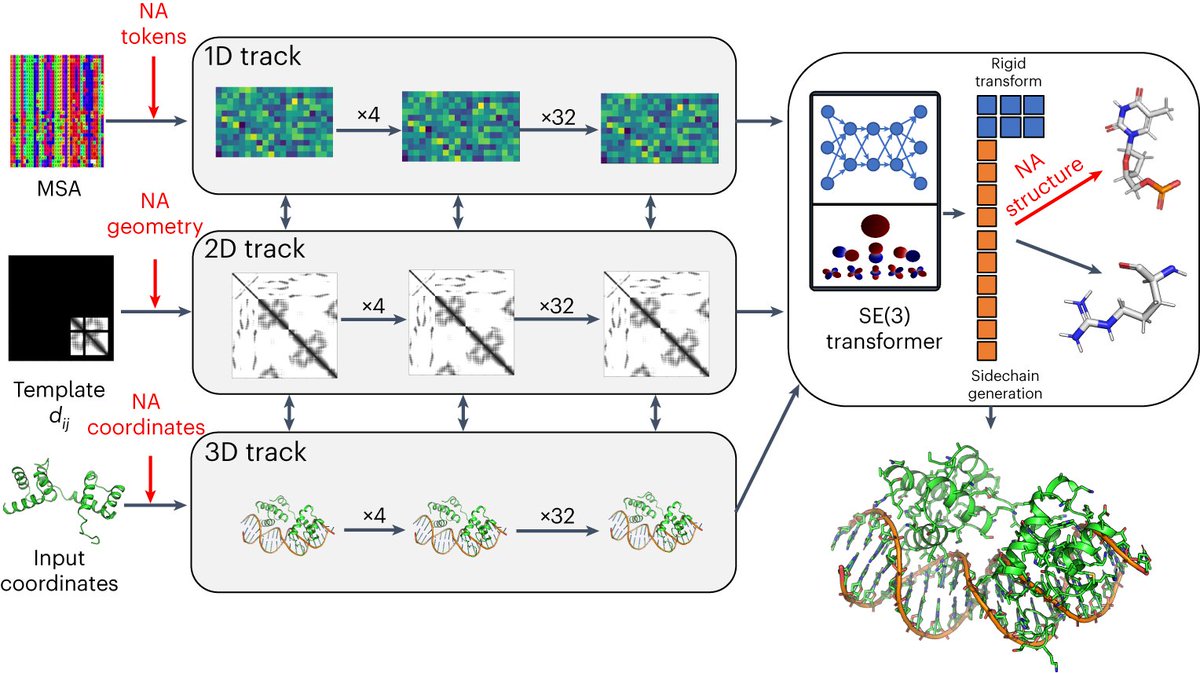
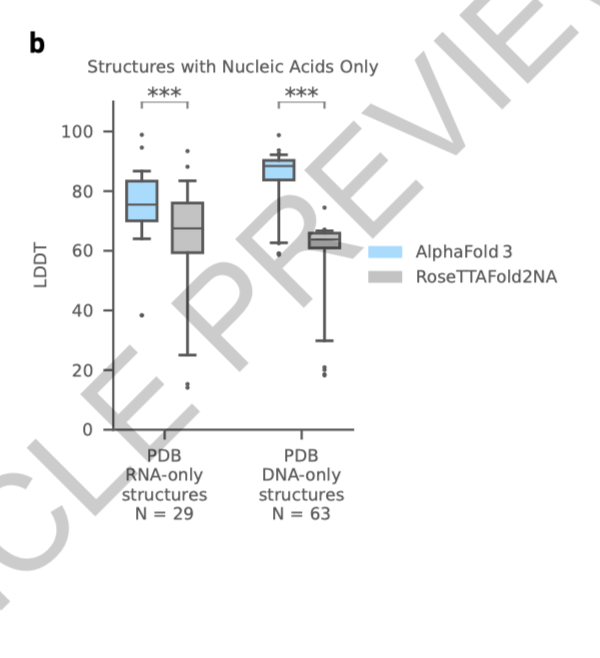
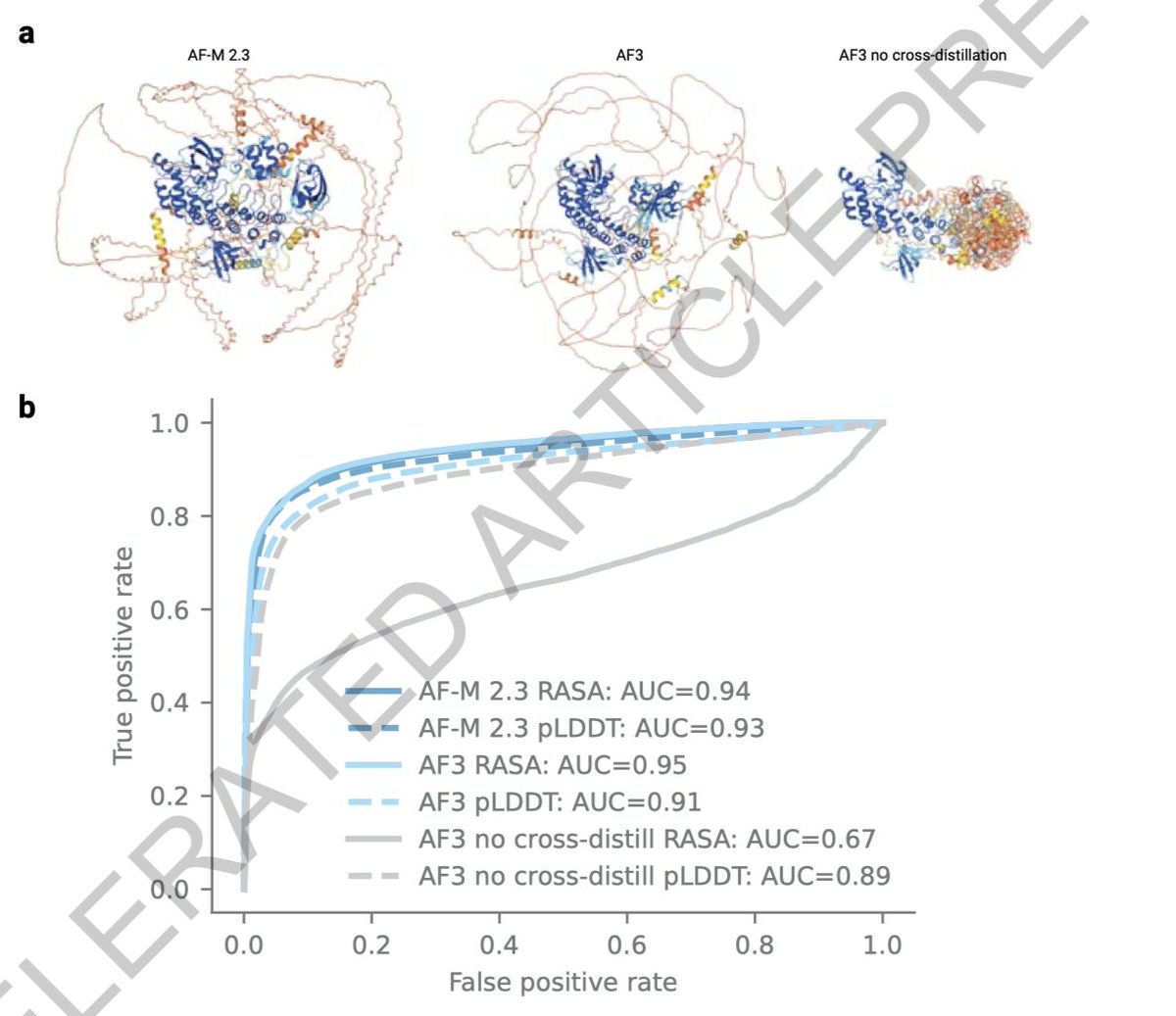
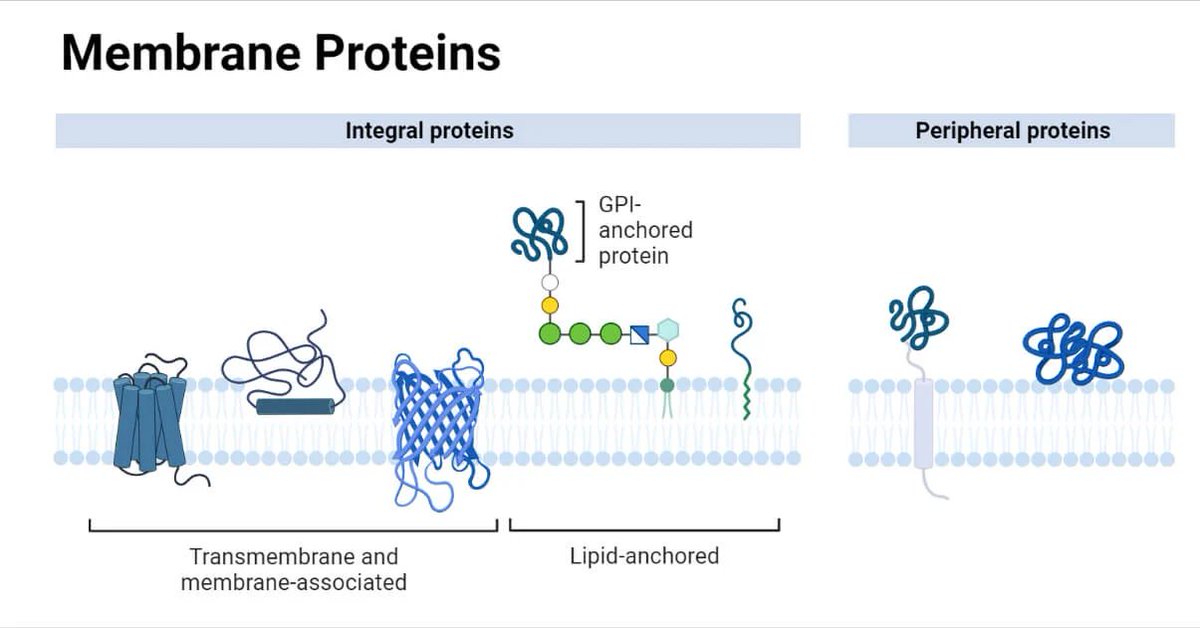
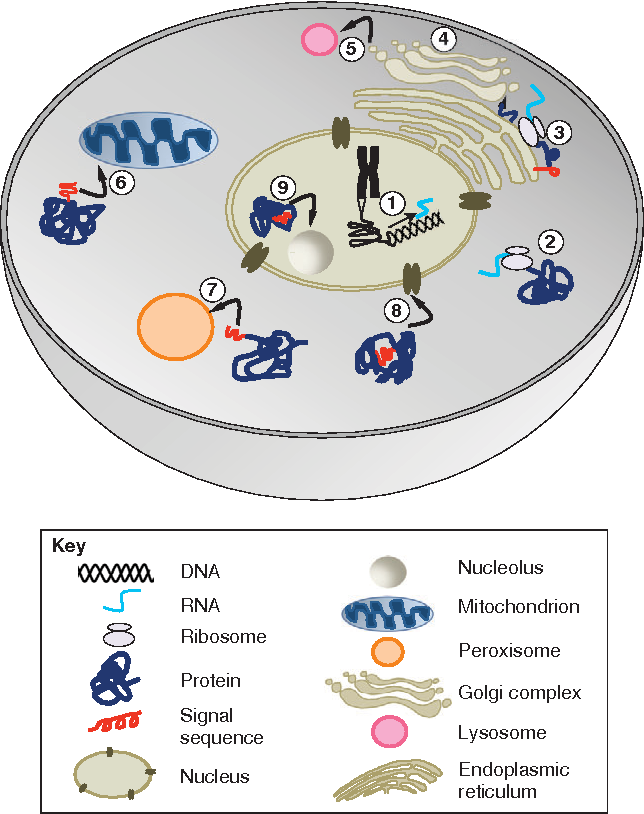
finally, computational challenges: the server limits exploration with a token ceiling of 5,000, affecting large systems like epigenetic complexes and full nuclear structures. Future advances must expand computational capacity to fully unravel these complex biological systems.
DeepMind's progress from AlphaFold 2 to AF3 illustrates the rapid evolution in using AI for biological insights. As we continue to refine these models, the potential for breakthroughs in understanding diseases and developing new therapies grows exponentially.
Loading suggestions...